GB Kinetics Example
This example demonstrates how to run the GB kinetics script and walks through where the results are stored. For this example, we focus on a \(\Sigma\) 17[001](410) STGB in Copper (Cu) with box length along the GB = 8 x GB period and box length along tilt axis = 4 x CSL length along tilt axis.
Open GB_kinetics.py and input Gb information, displacements recorded earlier and parameters for optimal transport calculations. Start with specifing the parameters required to identify the material and GB such as sigma number, misorientation, inclination, material symbol, and tilt axis.
size_along_gb_perioddetermines the length of the box along the gb period. The length of the box generated is 2*period*size_along_gb_period.size_along_tilt_axisdetermines the length of the box along the tilt axis. The length of the box generated is 2*(CSL length along tilt axis)*size_along_tilt_axis.lattive_Vectorsdetermine what crystal system is under consideration.
1#---------- Input parameters for GB in question ---------------#
2# Element
3element = "Cu"
4# Sigma value of Gb under consideration
5sigma = 29
6# Misorientation of the Gb
7misorientation = 43.6
8# Inclination of the GB
9inclination = 0.0
10# Lattice parameter of the element (try using the lat par corresponding to the potential you intend to use)
11latticeParameter = 3.615
12# Tilt axis of GB
13axis = [0, 0, 1]
14# Size of system along the GB period in terms of 2*CSL period
15size_along_gb_period = 10
16# Size of system along the tilt axis in terms of 2*CSL period
17size__along_tilt_axis = 1
18# Lattice Vectors for the crystal system (current implementation is tested for fcc only)
19lattice_vector = np.array([[0.5, 0.5, 0.0],
20 [0.0, 0.5, 0.5],
21 [0.5, 0.0,0.5]])
Next specify the parameters for Sinkhorn Algorithm run which determines the atomic trajectories in transformed region.
regularizationParameterdetermines the regularization for Sinkhorn algorithm. It can be loosely thought of as a measure of temperature. LowerregularizationParametermeans algorithm is more contrained and gives out only the smallest displacement maps. HigherregularizationParametermight give out multiple atomic mappings some of them might not be the lowest displacement.maximumIterationsdetermines the maximum number of iterations done by Sinkhorn Algorithm. If the algorithm does not converge (code will raise this error), increase this parameter.chooseDisconnectionis an optional argument which allows users to choose the disconnection mode if they want. SetchooseDisconnection = Trueif you want to choose the disconnection mode,choose_disconnection = Falsewill automatically create disconnection mode with smallest burgers vector and corresponding step height.
1#-------------- Parameters for min-shuffle algorithm --------------#
2# Regularization parameter for min-shuffle algorithm
3regularizationParameter = 0.005
4# Maximum iterations for min-shuffle algorithm
5maximumIterations = 1000000
6# Put True if you want to choose the disconnection mode, False will automatically create disconnection mode with
7# smallest burgers vector and corresponding step height
8chooseDisconnection = True
Specify the file that contains bicrystallography data from oilab output in
oilab_output_file. Current implementation has output files included for 3 tilt axes ([001],[110],[111]), stored inDatafolder.output_folderis the folder in which the output of this program is stored.
1# ------------------------- Input and output folders -------------------- #
2# Location of bicrystallographic data obtained using oILAB
3oilab_output_file = "data/fcc0-10.txt"
4# Location of directory where output is to be stored, the program creates subdirectories
5# for each element, sigma value , misorientation and disconnection mode within it
6output_folder = "output/"
Specify locations of external programs needed for running this code. This includes location of lammps installation, mpi installation. Also indicate the full path to the lammps potential file you want to use.
1# ---------- Location of programs needed to run this (change these) ----------------------#
2# Location of directory where lmp_serial and lmp_mpi are stored
3lammps_location = "/opt/homebrew/bin"
4# Location of directory where mpirun is stored
5mpi_location = "/opt/homebrew/bin"
6# Full path to the potential to be used
7lammps_potential = "/opt/homebrew/Cellar/lammps/20240829-update1/share/lammps/potentials/Cu_mishin1.eam.alloy"
Input the displacements that minimize GB to minimum energy structure. Can be obtained using
gridSearch.pyscript.
1# Displacements obtained from running grid_search script
2disp_along_gb = -0.1
3disp_along_tilt = -0.9
Specify the parameters for NEB run.
partitionsis the linearly interpolated images used in NEB run.neb_modelets you choose if you want to run intermediate images through NEB or not.neb_mode= 1 -> NEB with intermediate images,neb_mode= 0 -> NEb with just the initial and final GB images
1# Parameters for neb run on lammps
2# Number of partitions used for neb calculations
3partitions = 4
4# Variable lets you choose if you want to run intermediate images through NEB or not.
5# mode = 1 -> NEB with intermediate images, mode = 0 -> NEb with just the initial and final GB images
6neb_mode = 1
7# Variable which allows for switching off automatically triggering neb calculations (in case you only need disconnection images)
8run_neb = False
Finally call the function that calls the required functions and carries out the procedure detailed in the paper.
1# Run
2if __name__ == "__main__":
3 results_folder_path = runGBkinetics(sigma,misorientation,inclination,latticeParameter,lattice_vector,
4 axis,size_along_gb_period,size__along_tilt_axis,element,
5 regularizationParameter,maximumIterations,
6 lammps_location,mpi_location,output_folder,lammps_potential,
7 disp_along_gb,disp_along_tilt,oilab_output_file,
8 chooseDisconnection,run_neb,neb_mode,partitions)
Save and run GB_kinetics.py
$ python3 GB_kinetics.py
User input
After running GB_kinetics.py, the program will show the input parameters pertinent to system
user wants to explore.
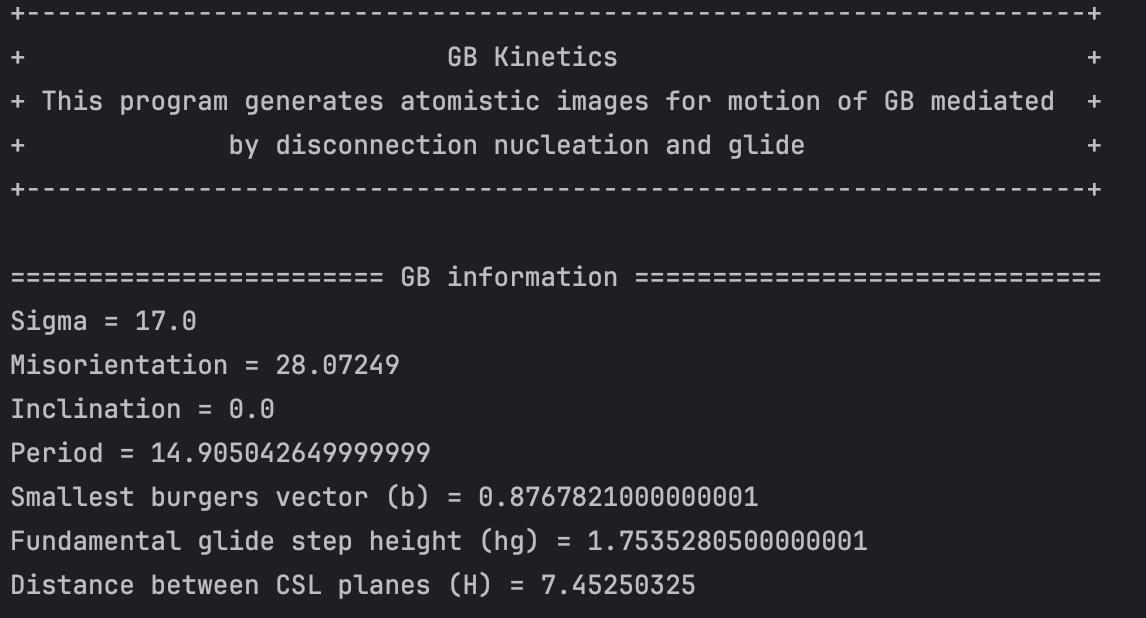
Then the program asks the user to choose the disconnection mode they want to construct. To do this, it lists out the disconnection modes calculated using GB information
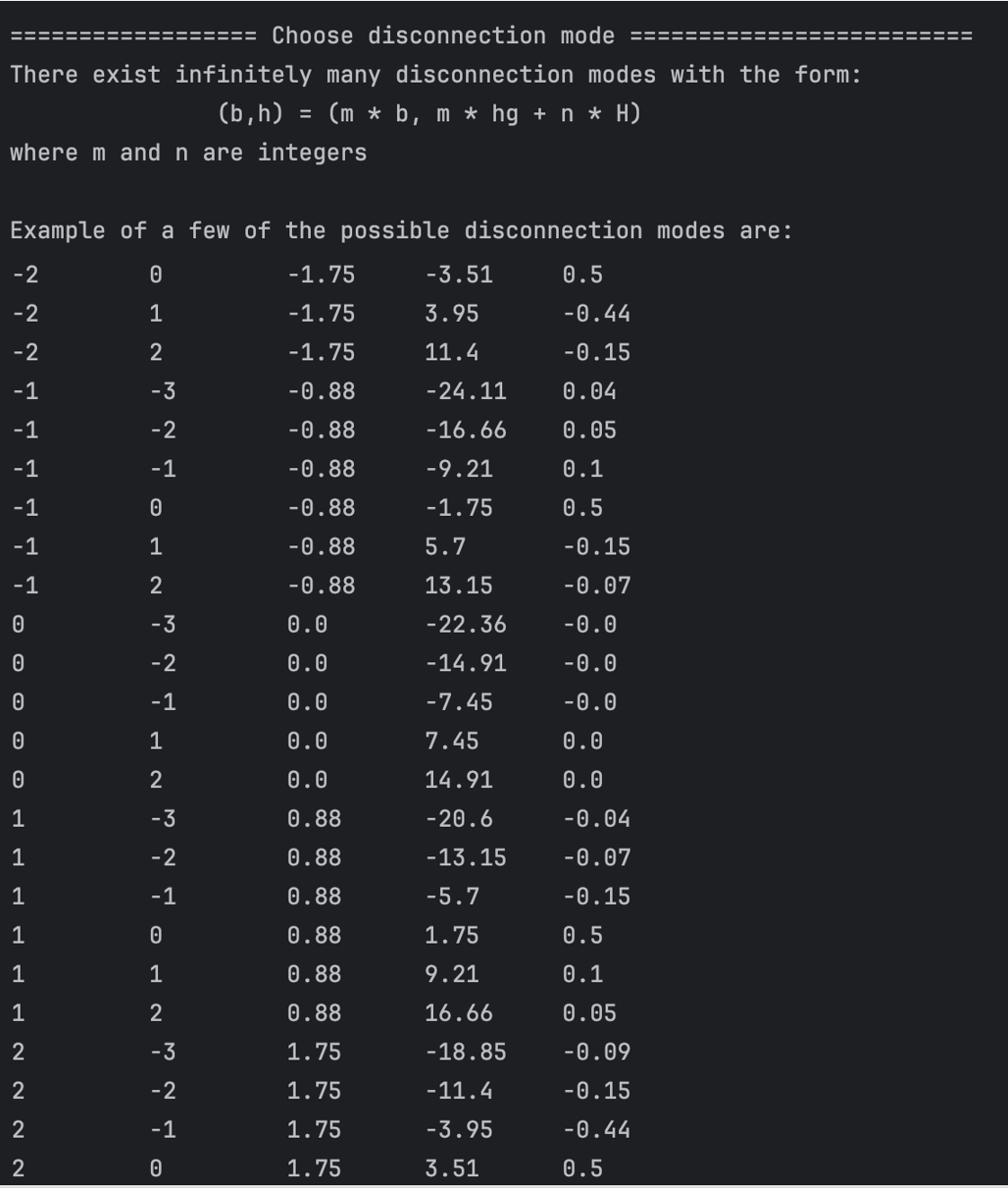
User can input the values of m and n, that corresponds to the disconnection mode they want to
construct. The program confirms the disconnection mode entered.
Command line Output
The code prints out which configuration it is generating and when completed, prints out the location of the output generated.
Output
All the generated configurations are stored in:
output/<Element>/Sigma<GB sigma number>/Misorientation<GB misorientation>.0/size<Size of box along GB>/b<Disconnection burgers vector>h<disconnection step height>
This code generates atomic configurations of different types:
LAMMPS data files for bicrystal with atoms obtained from bicrystallographic data and an applied plastic displacement , as discussed in Theory. These files are named as :
data.Cus<GB Sigma>.0inc<GB inclination>_size<Size along GB period>disc<configuration number>.
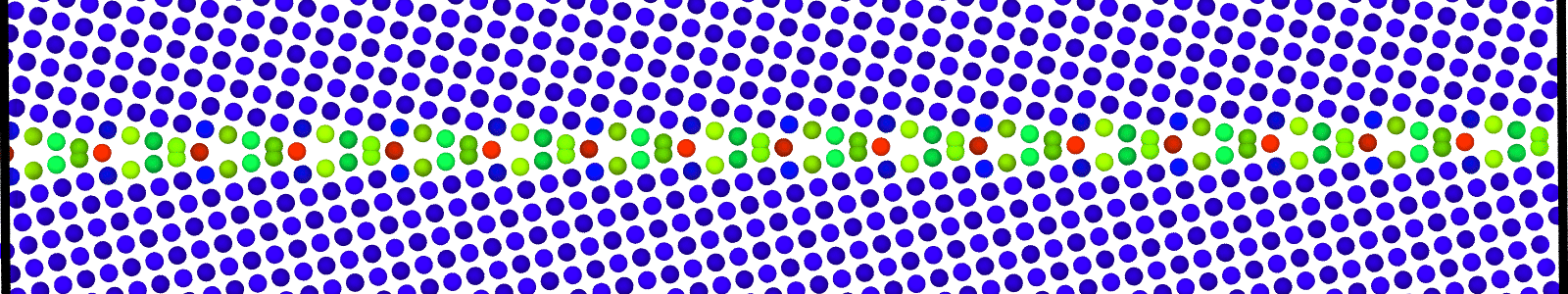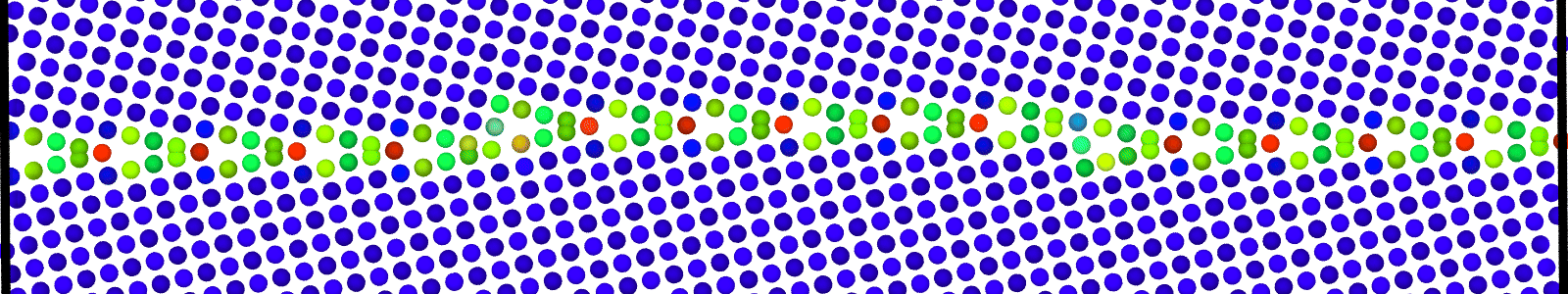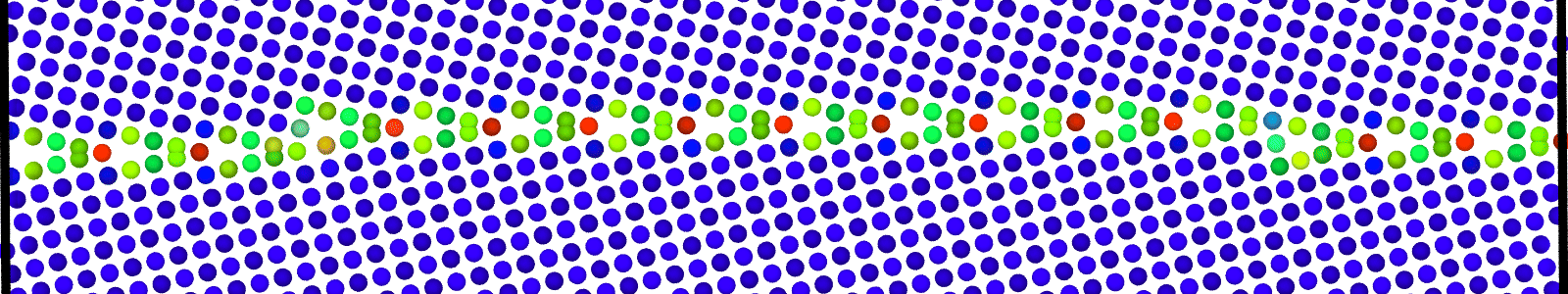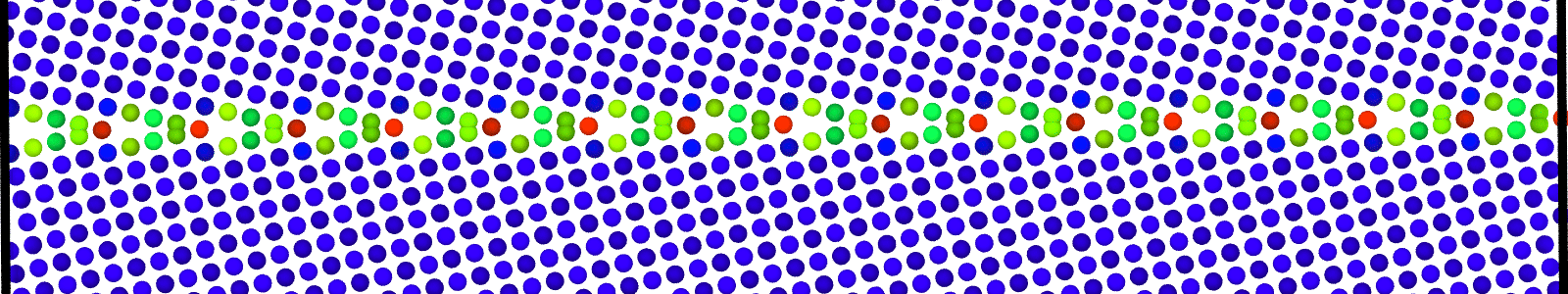
LAMMPS data files for minimized bicrystal with atoms now representing GB ground state. These files are named as :
data.Cus<GB Sigma>.0inc<GB inclination>_size<Size along GB period>disc<configuration number>_min.
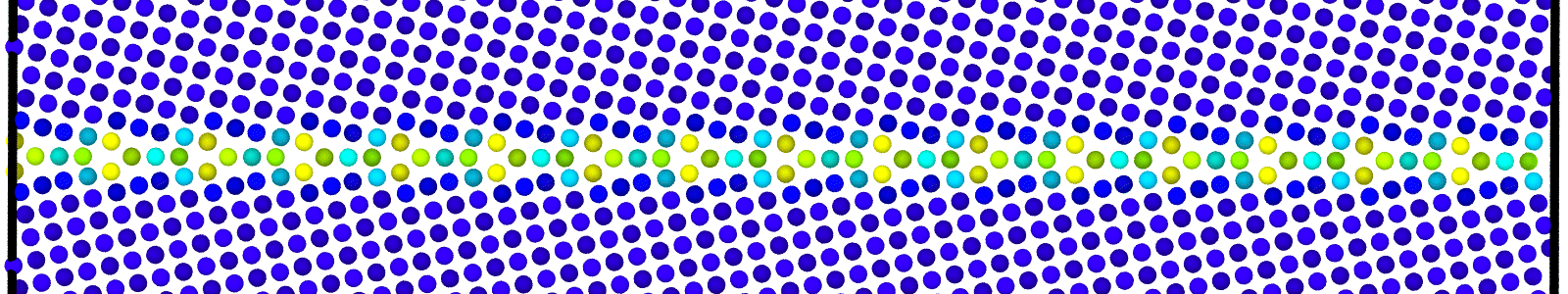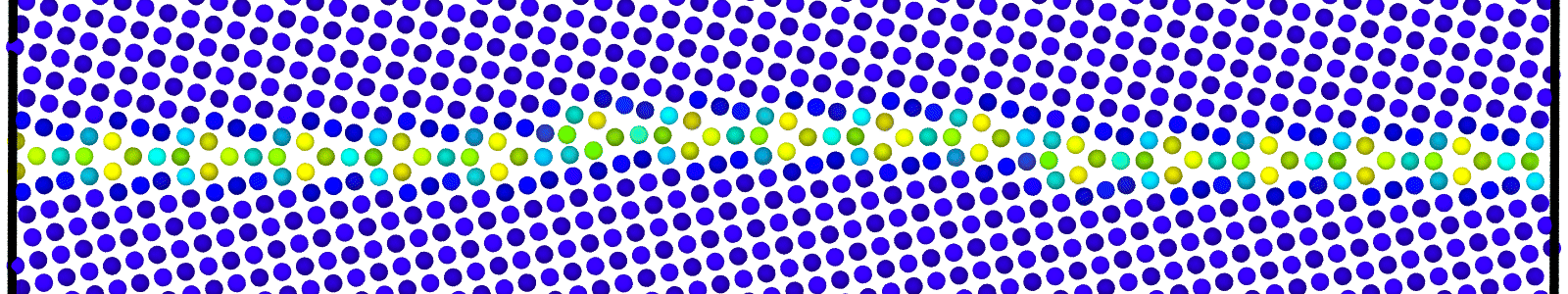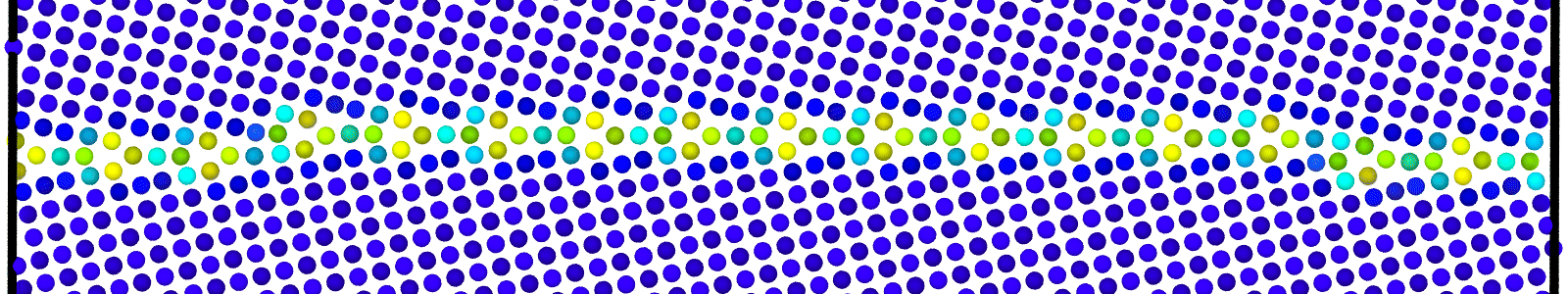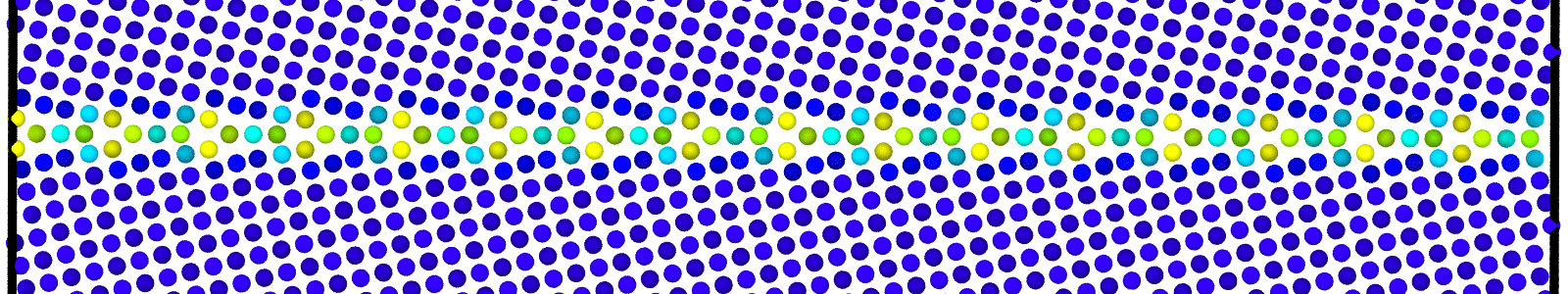
LAMMPS data files for bicrystals with atomic trajectories calculated using min-shuffle algorithm. These files are named as :
data.Cus<GB Sigma>.0inc<GB inclination>_size<Size along GB period>__step<configuration number>.
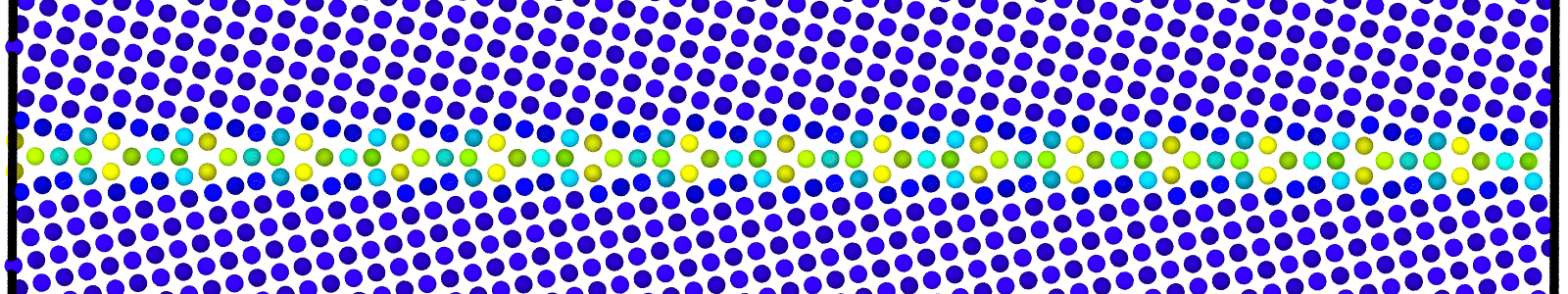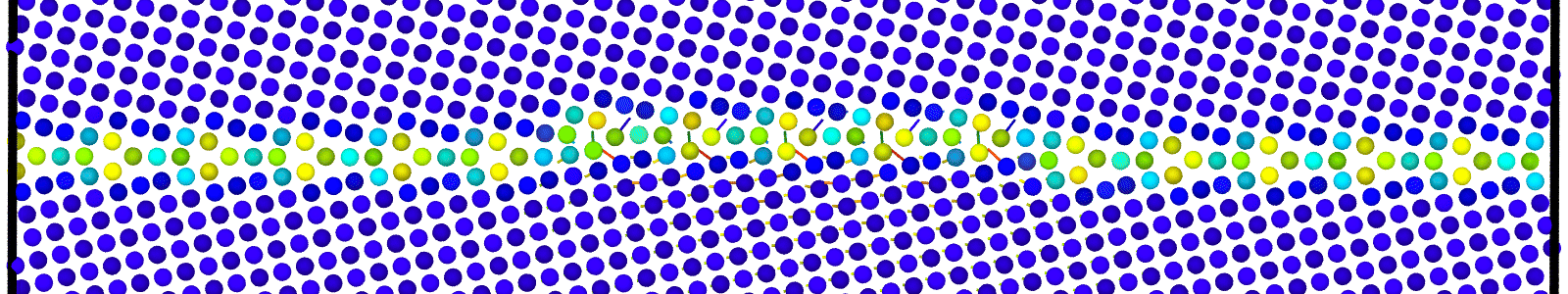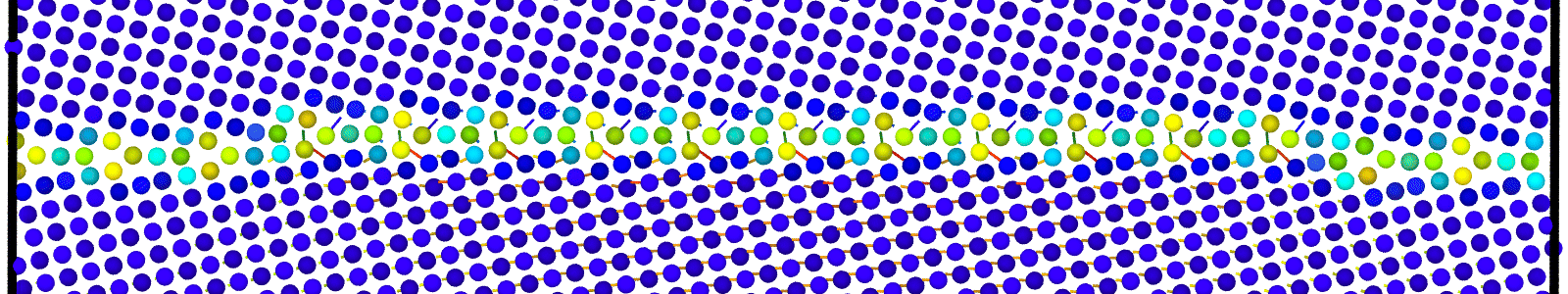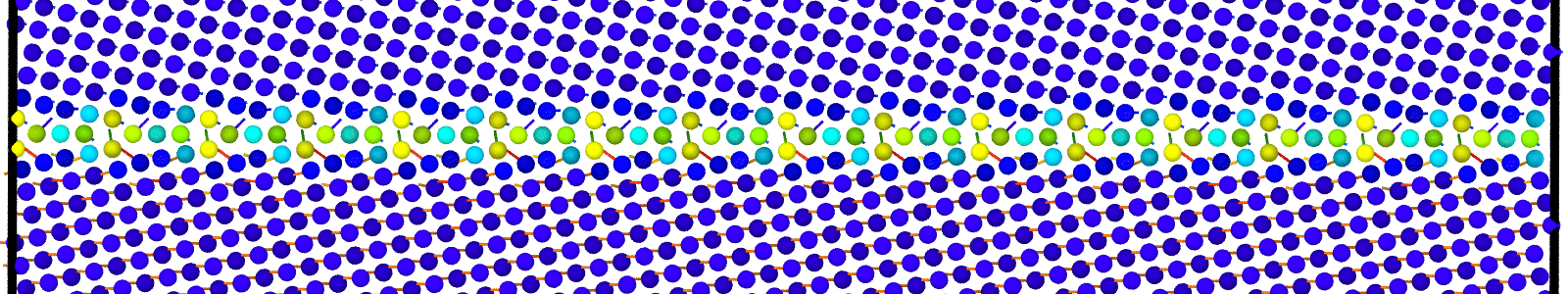
Data file corresponding to the bicrystals with atomic trajectories, containing the atoms that are to be included in
nebcalculations. These files are named as :data.Cus<GB Sigma>.0inc<GB inclination>_size<Size along GB period>_out_step<configuration number>.
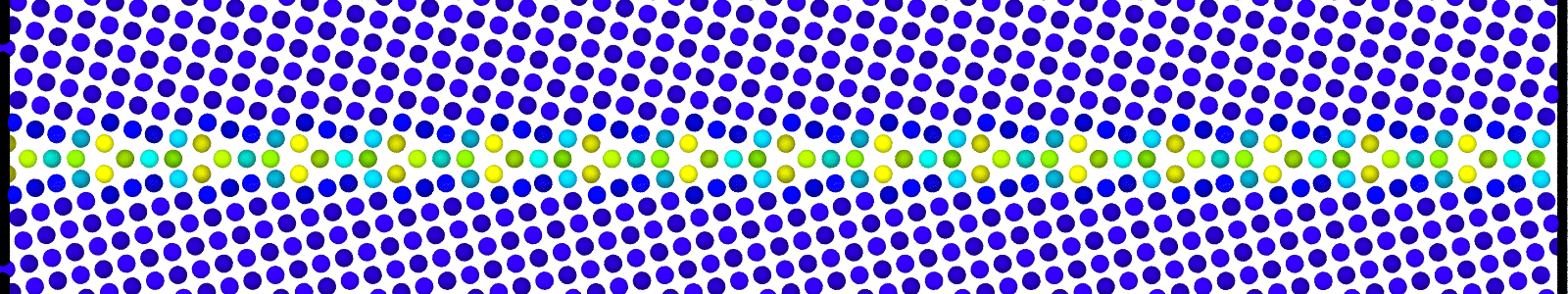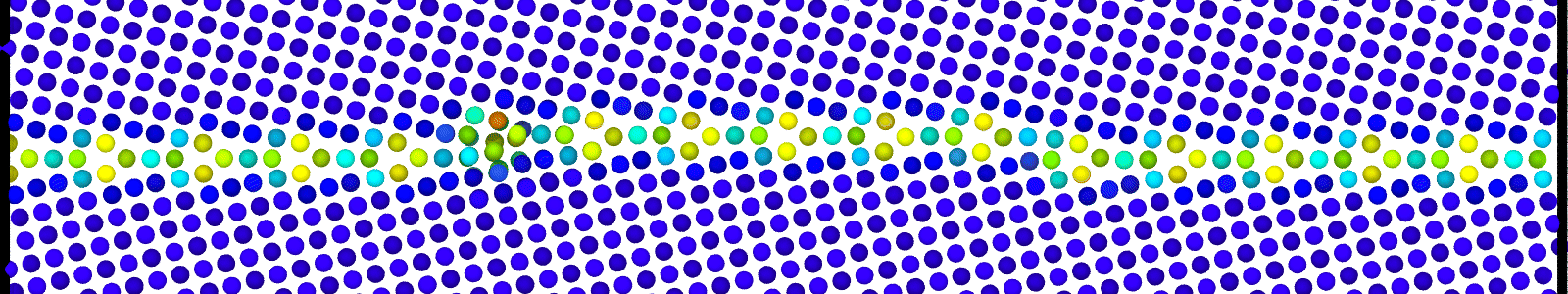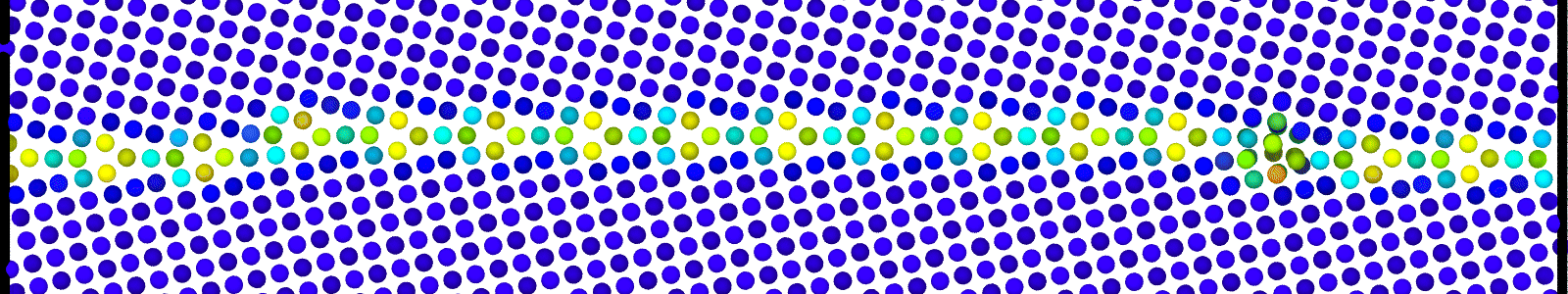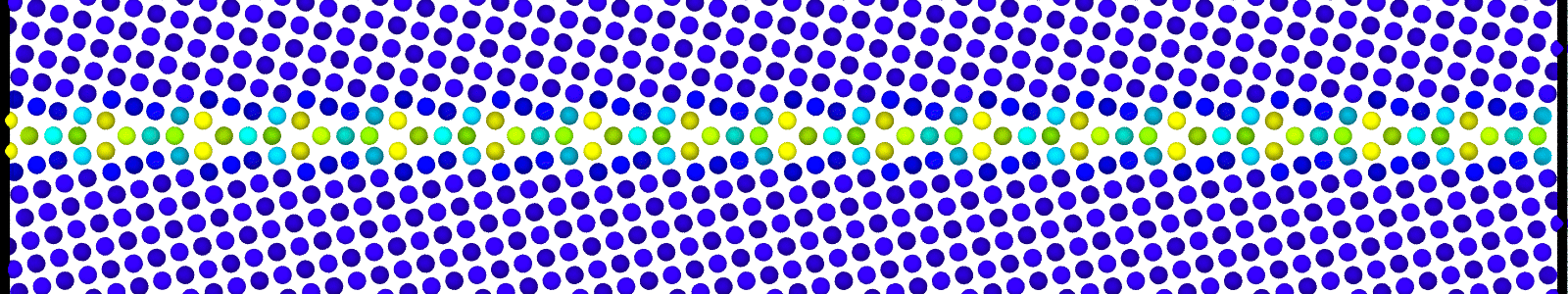
A LAMMPS data file containing multiple timesteps showing minimization of the bicrystal generated from bicrystallography. These files are named as :
data.Cus<GB Sigma>.0inc<GB inclination>_size<Size along GB period>disc<configuration number>_minmov.
The post processing function also plots the MEP w.r.t the reaction coordinates.Here, we present the results obtained by running this code and for comparison to existing method,we compare it with MEP calculated by Combe et. al. [1] for the same GB and box dimensions.
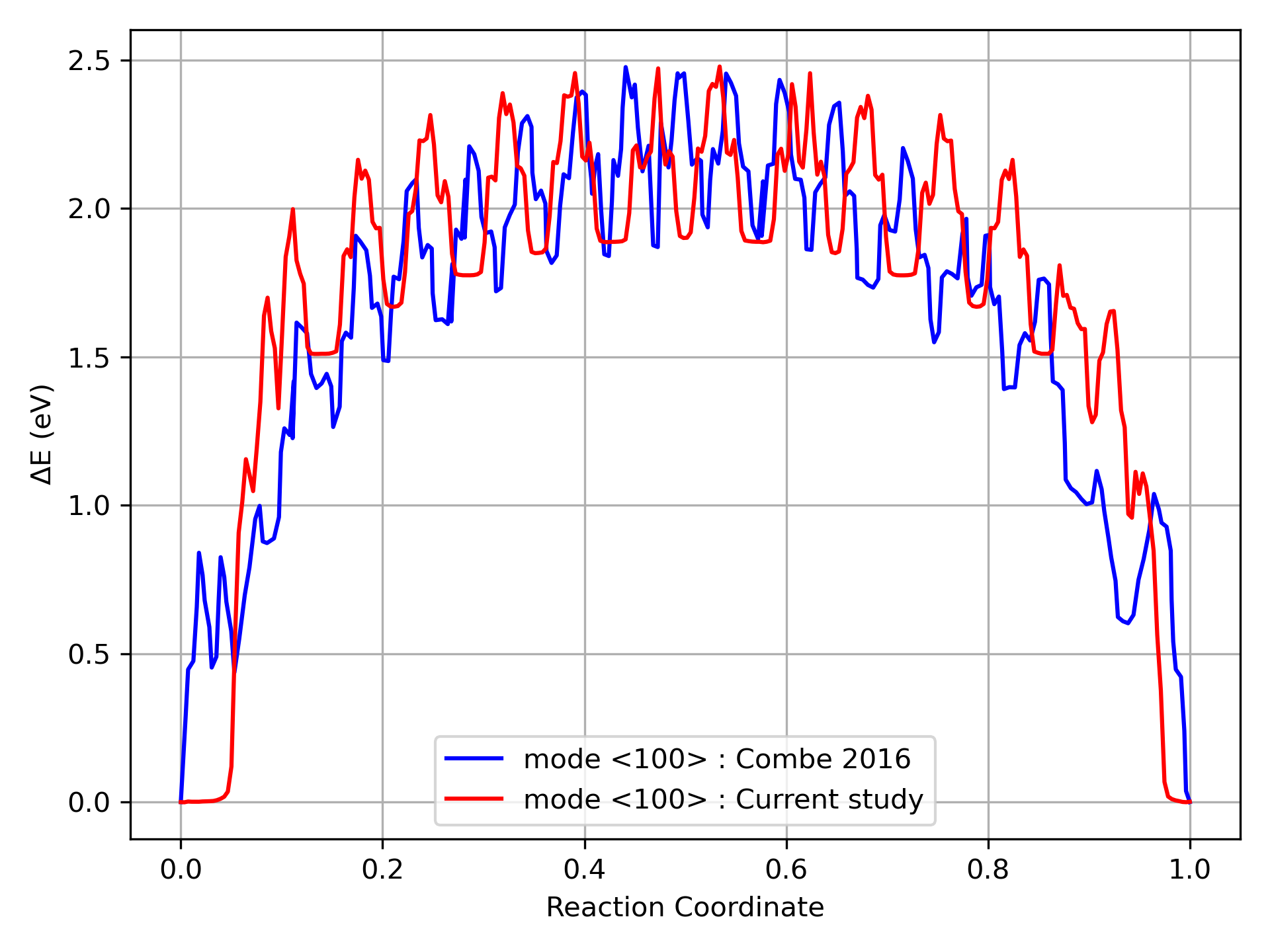
References
[1] N. Combe, F. Mompiou, M. Legros, Disconnections kinks and competing modes in shear-coupled grain boundary migration, Physical Review B 93 (2016) 024109.
Summary of important functions/classes used in the workflow
Bicrystals created in Construction of STGB microstate and Construction of atomistic GB migration images are objects of
src.bicrystal.bicrystalclass.The function
src.bicrystal.bicrystal.create_flat_gb_bicrystal()uses the methodology described in Construction of STGB microstate to create a bicrystal with as cut flat gb.The function
src.bicrystal.bicrystal.create_disconnection_containing_bicrystal()uses the methodology described in Construction of atomistic GB migration images to create a bicrystal with a disconnection step.The function
src.bicrsytal.bicrsytal.ordered_write()writes the bicrystal configurations generated intoLAMMPS data files.
Grid search required in Construction of STGB microstate which in turn utilizes utilizes
src.GBKineticsRunController.runGridSearch().GB bicrystallographic data is read from oILAB output in Data using
src.bicrystallography.bicrystallography.gb_props()All calls to
LAMMPSare handled by object of classsrc.runLAMMPS.run_LAMMPS.The function
src.runLAMMPS.run_LAMMPS.run_minimization()calls LAMMPS to minimize the bicrystal.The function
src.runLAMMPS.run_LAMMPS.run_grid_search()calls LAMMPS to run \(\gamma\) -surface method on a GB.The function
src.runLAMMPS.run_LAMMPS.run_neb_calc()calls LAMMPS to run a nudged elastic band calculation on the atomic configurations constructed in Construction of atomistic GB migration images.
Calculation of min-shuffle based atomic trajectories is done using an object of class
src.min_shuffle.min_shuffle.